trio,
gromacs and
Raster3D :
- Start from my.dcd and a PDB file (say, my.pdb) containing a set of atoms corresponding to those present in the DCD file (for example, from psfgen).
- Convert the DCD file to a format understood by gromacs :
trio my.dcd my.g96
- Run g_rmsf (from the gromacs suite) :
/usr/local/gromacs/i686-pc-linux-gnu/bin/g_rmsf -f my.g96 -s my.pdb -oq aniso.pdb -aniso
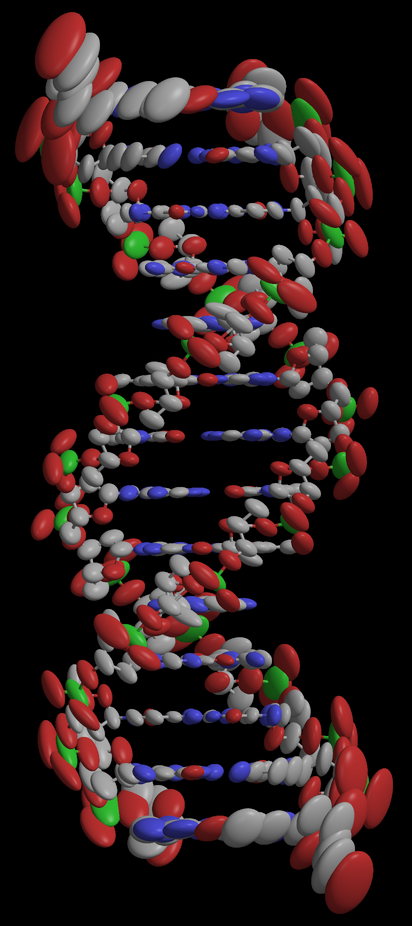
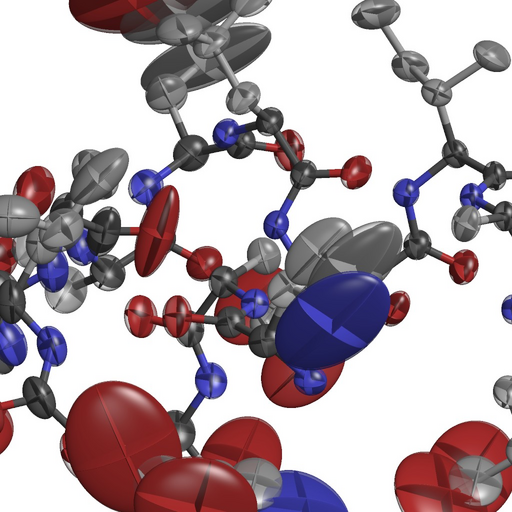
- If all goes well, try to make a picture of it :
rastep -nohydrogens -fancy3 < aniso.pdb | render -size 1024x1024 -zoom 1.5 -jpeg picture.jpg ; display picture.jpg
![[Home]](/wiki.png)