If you are lucky enough to have a NMR structure deposited with the PDB for your molecule, you can make an interesting comparison between the variance-covariance matrix obtained from the NMR ensemble and the matrix obtained from the molecular dynamics simulation.
Obtaining the covariance matrix from the MD run has been discussed in detail (see Variance-covariance and cross-correlation analysis). The difficult part is obtaining the covariance matrix from the ensemble of NMR stuctures. The important thing to notice is that by its definition the covariance matrix
(x_j20-203cx_j3e)3e0a202424.png)
is time-independent. This means that the order of structures is irrelevant (the same matrix will be obtained from the same structures no matter what their order is). The trick then is to prepare a pseudo-trajectory DCD file containing the NMR structures and a PSF file for them. Then you use
carma in the usual way to prepare the matrix. Converting a series of PDB files to a DCD file can easily be done with VMD (select molecule, then 'Save coordinates' and chose DCD). The resulting DCD file (from VMD) must be filtered through
catdcd
catdcd -o mynew.dcd previous.dcd
Then run carma as before and thats it. You can now compare the two matrices (the example shown below is for native Rop) :
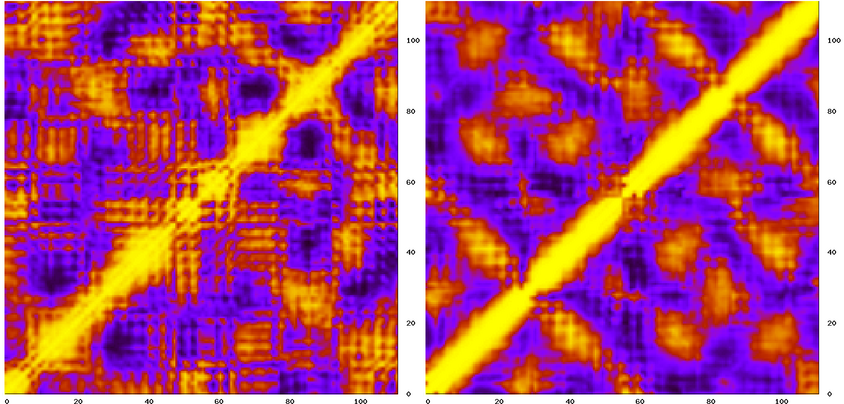
The peaky appearance of the NMR-derived map (on the left) is possibly due to the small number of structures (10 in this case) that were used for the calculation.
(x_j20-203cx_j3e)3e0a202424.png)
![[Home]](/wiki.png)