- Create a clean directory and cd in it.
- Get the CHARMM force field parameter and topology files :
- cp /usr/local/charmm27/par_all27_prot_na.inp ./
- cp /usr/local/charmm27/top_all27_prot_na.inp ./
- Download the coordinates for Dickerson's dodecamer from PDB [1] and save it on your disk.
- gunzip 1rop.pdb1.gz
- mv 1rop.pdb1 dimer.pdb
- Edit the file dimer.pdb and remove all lines that do not start with ATOM or TER. Also, retain the final END. Now the difficult part : although the file contains two chains, these have the same identifier ' A '. You should find the beginning of the second chain and then use find-and-replace (or use a macro) to change only for the second chain all occurences of the chain identifier ' A ' to ' B '. Save the modified file which should look like this :
ATOM 1 N MET A 1 31.007 2.290 18.034 1.00 28.97 N
ATOM 2 CA MET A 1 32.390 2.582 17.546 1.00 26.82 C
ATOM 3 C MET A 1 32.808 1.432 16.618 1.00 21.98 C
ATOM 4 O MET A 1 32.375 0.280 16.846 1.00 23.60 O
......................
ATOM 445 CE1 PHE A 56 38.342 8.335 16.790 1.00 25.49 C
ATOM 446 CE2 PHE A 56 39.530 7.284 18.653 1.00 23.77 C
ATOM 447 CZ PHE A 56 39.527 7.962 17.421 1.00 24.84 C
TER 448 PHE A 56
ATOM 1 N MET B 1 74.680 2.290 35.339 1.00 28.97 N
ATOM 2 CA MET B 1 73.297 2.582 35.827 1.00 26.82 C
ATOM 3 C MET B 1 72.879 1.432 36.755 1.00 21.98 C
ATOM 4 O MET B 1 73.312 0.280 36.526 1.00 23.60 O
......................
ATOM 446 CE2 PHE B 56 66.157 7.284 34.719 1.00 23.77 C
ATOM 447 CZ PHE B 56 66.160 7.962 35.952 1.00 24.84 C
TER 448 PHE B 56
END
- rasmol dimer.pdb (you should see the Rop dimer).
- Now it is time to align the axes of inertia of the molecule with the orthogonal frame : prepare a file with the name moleman.csh containing the following :
- Run it : source moleman.csh. This should create two files : a log file (moleman.log) and the new pdb (aligned.pdb).
- rasmol aligned.pdb (to confirm the changes)
- We need to prepare two pdb files each containing the coordinates of each of the two chains. The simplest approach is :
- cp aligned.pdb A.pdb
- cp aligned.pdb B.pdb
- Edit A.pdb and retain only the A-chain coordinates. Add an END at the end of the file. Find-and-replace 'HIS' with 'HSP' (fully protonated histidines, corresponding to performing a run at pH < 6). Save the file. Repeat for B.pdb. For example, A.pdb should look like this :
ATOM 1 N MET A 1 22.545 -6.589 6.155 1.00 28.97 N
ATOM 2 CA MET A 1 21.754 -5.355 5.863 1.00 26.82 C
ATOM 3 C MET A 1 21.994 -4.366 7.013 1.00 21.98 C
......................
ATOM 445 CE1 PHE A 56 17.522 -1.101 0.110 1.00 25.49 C
ATOM 446 CE2 PHE A 56 15.440 -1.841 1.161 1.00 23.77 C
ATOM 447 CZ PHE A 56 16.199 -0.871 0.483 1.00 24.84 C
END
- Create a file with the name psfgen.csh containing the following :
- Use it to generate a new pdb file (psfgen.pdb) and the PSF file (psfgen.psf) needed for NAMD : source psfgen.csh. Examine the output from the program (file psfgen.log).
- Create a file with the name make_all.vmd containing the following script (for VMD) :
- Run this script via VMD :
- Start VMD : vmd
- Locate VMD's console (window with the prompt)
- In the console give source make_all.vmd
- If all goes well you should see your fully hydrated and ionised system on VMD's graphics window.
- To finish type quit in the console.
- The last step before running NAMD is to determine the limits (along the orthogonal frame) of the system. A fast method to do this is to use the program pdbset from the CCP4 suite of program. Here it is how :
- Start it : pdbset xyzin ionized.pdb >& pdbset.log
- Type END
- Edit the file pdbset.log. Somewhere near the end of it you will see something similar to :
Orthogonal Coordinate limits in output file:
Minimum Maximum Centre Range
On X : -39.30 39.30 0.00 78.60
On Y : -30.05 30.05 0.00 60.10
On Z : -25.98 30.85 2.44 56.83
Note these numbers, especially the range (on x,y,z) and position of centre.
- Create a NAMD minimisation and heating-up script with the name heat.namd containing the following :
- You can now give it a quick try with NAMD just to make sure it starts without problems. To avoid overloading machines that are already busy with real calculations you must review the cluster usage as follows :
- Type mosmon from your terminal. You should see something similar to this
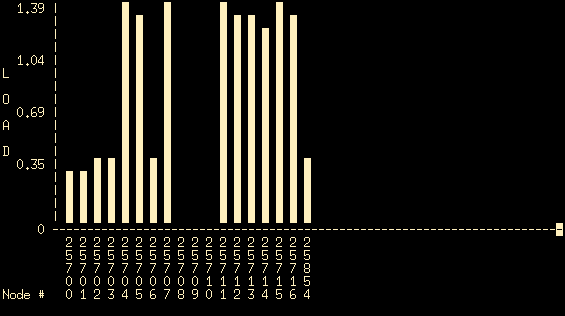 This graph shows (in real time) the CPU load for each of the nodes that are currently part of the cluster. The horizontal axis shows the various nodes, the vertical axis shows the corresponding load. If all machines are working properly and they are all running the openMosix linux kernel you should see at least 18 nodes (numbered 25700-25716, plus another node with ID 25854). In this case all cluster nodes are running a proper operating system (and can be seen in the mosmon output) and three of these (25708, 25709 and 25710 corresponding to pc08, pc09 and pc10) are idle. So, nodes pc08, pc09 and pc10 are good candidates for performing your tests.
This graph shows (in real time) the CPU load for each of the nodes that are currently part of the cluster. The horizontal axis shows the various nodes, the vertical axis shows the corresponding load. If all machines are working properly and they are all running the openMosix linux kernel you should see at least 18 nodes (numbered 25700-25716, plus another node with ID 25854). In this case all cluster nodes are running a proper operating system (and can be seen in the mosmon output) and three of these (25708, 25709 and 25710 corresponding to pc08, pc09 and pc10) are idle. So, nodes pc08, pc09 and pc10 are good candidates for performing your tests. - Type q to stop mosmon and then give qmon (from your terminal). The main Sun Grid Engine GUI window should appear:
 Select "Queue control". This should pop-up the corresponding window :
Select "Queue control". This should pop-up the corresponding window : 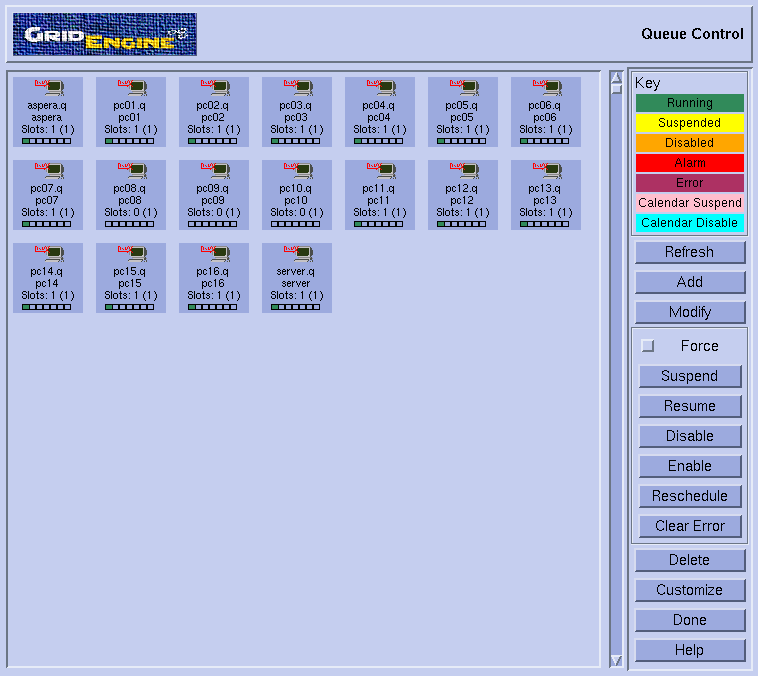 Note the small green boxes (indicating that a job is running) on all nodes except pc08, pc09 and pc10. So, it does look like we could use these three machines for the tests. Press 'Done' on the queue control window and then exit in the qmon window.
Note the small green boxes (indicating that a job is running) on all nodes except pc08, pc09 and pc10. So, it does look like we could use these three machines for the tests. Press 'Done' on the queue control window and then exit in the qmon window.
- As a first test lets run NAMD on a single node, e.g. pc09 : login to it with ssh pc9 (use your password to login). Then cd to the directory containing your file : oops, no such file or directory ? That's right : you created the files on your local machine, which probably isn't pc09. So, here comes the need for a cluster-wide filesystem. This is implemented in the form of the /work directory. Proceed as follows :
- Logout from pc09 : exit
- cd /work
- ls
- cd tmp
- mkdir my_tests
- Now go back to the directory containing the files you created. Of all the files present in there you only need to copy those few :
cp ionized.p* par_all27_prot_na.inp heat.namd fix_backbone.pdb restrain_ca.pdb /work/tmp/my_tests/
- ssh pc9 and login again
- cd /work/tmp/my_tests/
- ls and you should see your files (although you are connected to pc9).
- mkdir output will create a subdirectory where the output files from NAMD will be written (during the run).
- Now, go for it : runhome /usr/local/NAMD_2.5/namd2 heat.namd. The runhome command tells the openMosix kernel to run the job on the local machine (not to migrate it away). Once the program performs a couple of minimisation steps stop it with <CTRL-C>. What you should see should be similar to :
Charm++: standalone mode (not using charmrun)
Info: NAMD 2.5 for Linux-i686
Info:
Info: Please visit http://www.ks.uiuc.edu/Research/namd/
Info: and send feedback or bug reports to namd@ks.uiuc.edu
Info:
Info: Please cite Kale et al., J. Comp. Phys. 151:283-312 (1999)
Info: in all publications reporting results obtained with NAMD.
Info:
Info: Based on Charm++/Converse 050612 for net-linux-icc
Info: Built Fri Sep 26 17:33:59 CDT 2003 by jim on lisboa.ks.uiuc.edu
Info: Sending usage information to NAMD developers via UDP. Sent data is:
Info: 1 NAMD 2.5 Linux-i686 1 pc09.cluster.mbg.gr glykos
Info: Running on 1 processors.
Info: 1335 kB of memory in use.
Measuring processor speeds... Done.
Info: Configuration file is heat.namd
TCL: Suspending until startup complete.
Info: SIMULATION PARAMETERS:
Info: TIMESTEP 2
Info: NUMBER OF STEPS 0
Info: STEPS PER CYCLE 8
Info: PERIODIC CELL BASIS 1 79.6 0 0
Info: PERIODIC CELL BASIS 2 0 61.1 0
Info: PERIODIC CELL BASIS 3 0 0 57.83
Info: PERIODIC CELL CENTER 0 0 2.44
Info: WRAPPING WATERS AROUND PERIODIC BOUNDARIES ON OUTPUT.
Info: WRAPPING TO IMAGE NEAREST TO PERIODIC CELL CENTER.
Info: LOAD BALANCE STRATEGY Other
Info: LDB PERIOD 1600 steps
Info: FIRST LDB TIMESTEP 40
Info: LDB BACKGROUND SCALING 1
Info: HOM BACKGROUND SCALING 1
Info: PME BACKGROUND SCALING 1
Info: MAX SELF PARTITIONS 50
Info: MAX PAIR PARTITIONS 20
Info: SELF PARTITION ATOMS 125
Info: PAIR PARTITION ATOMS 200
Info: PAIR2 PARTITION ATOMS 400
Info: INITIAL TEMPERATURE 0
Info: CENTER OF MASS MOVING? NO
Info: DIELECTRIC 1
Info: EXCLUDE SCALED ONE-FOUR
Info: 1-4 SCALE FACTOR 1
Info: DCD FILENAME output/heat_out.dcd
Info: DCD FREQUENCY 200
Warning: INITIAL COORDINATES WILL NOT BE WRITTEN TO DCD FILE
Info: XST FILENAME output/heat_out.xst
Info: XST FREQUENCY 200
Info: NO VELOCITY DCD OUTPUT
Info: OUTPUT FILENAME output/heat_out
Info: RESTART FILENAME output/restart
Info: RESTART FREQUENCY 1000
Info: BINARY RESTART FILES WILL BE USED
Info: SWITCHING ACTIVE
Info: SWITCHING ON 10
Info: SWITCHING OFF 12
Info: PAIRLIST DISTANCE 13.5
Info: PAIRLIST SHRINK RATE 0.01
Info: PAIRLIST GROW RATE 0.01
Info: PAIRLIST TRIGGER 0.3
Info: PAIRLISTS PER CYCLE 2
Info: PAIRLISTS ENABLED
Info: MARGIN 0.48
Info: HYDROGEN GROUP CUTOFF 2.5
Info: PATCH DIMENSION 16.48
Info: ENERGY OUTPUT STEPS 20
Info: TIMING OUTPUT STEPS 200
Info: FIXED ATOMS ACTIVE
Info: FORCES BETWEEN FIXED ATOMS ARE CALCULATED
Info: HARMONIC CONSTRAINTS ACTIVE
Info: HARMONIC CONS EXP 2
Info: LANGEVIN DYNAMICS ACTIVE
Info: LANGEVIN TEMPERATURE 320
Info: LANGEVIN DAMPING COEFFICIENT IS 10 INVERSE PS
Info: LANGEVIN DYNAMICS APPLIED TO HYDROGENS
Info: LANGEVIN PISTON PRESSURE CONTROL ACTIVE
Info: TARGET PRESSURE IS 1.01325 BAR
Info: OSCILLATION PERIOD IS 200 FS
Info: DECAY TIME IS 100 FS
Info: PISTON TEMPERATURE IS 320 K
Info: PRESSURE CONTROL IS GROUP-BASED
Info: INITIAL STRAIN RATE IS 0 0 0
Info: CELL FLUCTUATION IS ISOTROPIC
Info: PARTICLE MESH EWALD (PME) ACTIVE
Info: PME TOLERANCE 1e-06
Info: PME EWALD COEFFICIENT 0.257952
Info: PME INTERPOLATION ORDER 4
Info: PME GRID DIMENSIONS 80 64 60
Info: Attempting to read FFTW data from FFTW_NAMD_2.5_Linux-i686.txt
Info: Optimizing 6 FFT steps. 1... 2... 3... 4... 5... 6... Done.
Info: Writing FFTW data to FFTW_NAMD_2.5_Linux-i686.txt
Info: FULL ELECTROSTATIC EVALUATION FREQUENCY 4
Info: USING VERLET I (r-RESPA) MTS SCHEME.
Info: C1 SPLITTING OF LONG RANGE ELECTROSTATICS
Info: PLACING ATOMS IN PATCHES BY HYDROGEN GROUPS
Info: RIGID BONDS TO HYDROGEN : ALL
Info: ERROR TOLERANCE : 1e-08
Info: MAX ITERATIONS : 100
Info: RIGID WATER USING SETTLE ALGORITHM
Info: NONBONDED FORCES EVALUATED EVERY 2 STEPS
Info: RANDOM NUMBER SEED 1099129124
Info: USE HYDROGEN BONDS? NO
Info: COORDINATE PDB ionized.pdb
Info: STRUCTURE FILE ionized.psf
Info: PARAMETER file: CHARMM format!
Info: PARAMETERS par_all27_prot_na.inp
Info: SUMMARY OF PARAMETERS:
Info: 250 BONDS
Info: 622 ANGLES
Info: 1049 DIHEDRAL
Info: 73 IMPROPER
Info: 130 VDW
Info: 0 VDW_PAIRS
Info: ****************************
Info: STRUCTURE SUMMARY:
Info: 25368 ATOMS
Info: 17518 BONDS
Info: 11126 ANGLES
Info: 4764 DIHEDRALS
Info: 308 IMPROPERS
Info: 0 EXCLUSIONS
Info: 112 CONSTRAINTS
Info: 450 FIXED ATOMS
Info: 24450 RIGID BONDS
Info: 0 RIGID BONDS BETWEEN FIXED ATOMS
Info: 50304 DEGREES OF FREEDOM
Info: 8768 HYDROGEN GROUPS
Info: 226 HYDROGEN GROUPS WITH ALL ATOMS FIXED
Info: TOTAL MASS = 154680 amu
Info: TOTAL CHARGE = 1.60187e-06 e
Info: *****************************
Info: Entering startup phase 0 with 7505 kB of memory in use.
Info: Entering startup phase 1 with 7505 kB of memory in use.
Info: Entering startup phase 2 with 9341 kB of memory in use.
Info: Entering startup phase 3 with 9541 kB of memory in use.
Info: PATCH GRID IS 4 (PERIODIC) BY 3 (PERIODIC) BY 3 (PERIODIC)
Info: REMOVING COM VELOCITY 0 0 0
Info: LARGEST PATCH (17) HAS 822 ATOMS
Info: Entering startup phase 4 with 12754 kB of memory in use.
Info: PME using 1 and 1 processors for FFT and reciprocal sum.
Creating Strategy 4
Creating Strategy 4
Info: PME GRID LOCATIONS: 0
Info: PME TRANS LOCATIONS: 0
Info: Optimizing 4 FFT steps. 1... 2... 3... 4... Done.
Info: Entering startup phase 5 with 14006 kB of memory in use.
Info: Entering startup phase 6 with 14006 kB of memory in use.
Info: Entering startup phase 7 with 14006 kB of memory in use.
Info: COULOMB TABLE R-SQUARED SPACING: 0.0625
Info: COULOMB TABLE SIZE: 769 POINTS
Info: NONZERO IMPRECISION IN COULOMB TABLE: 2.64698e-22 (712) 2.64698e-22 (712)
Info: NONZERO IMPRECISION IN COULOMB TABLE: 1.26218e-29 (742) 1.57772e-29 (742)
Info: NONZERO IMPRECISION IN COULOMB TABLE: 1.27055e-21 (767) 1.48231e-21 (767)
Info: Entering startup phase 8 with 17392 kB of memory in use.
Info: Finished startup with 22280 kB of memory in use.
TCL: Minimizing for 0 steps
ETITLE: TS BOND ANGLE DIHED IMPRP ELECT VDW BOUNDARY MISC KINETIC TOTAL TEMP TOTAL2 TOTAL3 TEMPAVG PRESSURE GPRESSURE VOLUME PRESSAVG GPRESSAVG
ENERGY: 0 7954.7376 5748.6073 437.1674 41.0044 -82921.4480 14265.3625 0.0000 0.0000 0.0000 -54474.5687 0.0000 -54474.5687 -54474.5687 0.0000 5922.6194 7853.0443 281259.6748 5922.6194 7853.0443
TCL: Setting parameter langevinPiston to off
TCL: Minimizing for 400 steps
ETITLE: TS BOND ANGLE DIHED IMPRP ELECT VDW BOUNDARY MISC KINETIC TOTAL TEMP TOTAL2 TOTAL3 TEMPAVG PRESSURE GPRESSURE VOLUME PRESSAVG GPRESSAVG
ENERGY: 0 7954.7376 5748.6073 437.1674 41.0044 -82921.4480 14265.3625 0.0000 0.0000 0.0000 -54474.5687 0.0000 -54474.5687 -54474.5687 0.0000 5922.6194 7853.0443 281259.6748 5922.6194 7853.0443
INITIAL STEP: 1e-06
GRADIENT TOLERANCE: 28662.8
ENERGY: 1 7935.4448 5745.3453 437.0249 40.9796 -82926.1450 14017.9693 0.0000 0.0000 0.0000 -54749.3812 0.0000 -54749.3812 -54749.3812 0.0000 5666.8255 7600.7423 281259.6748 5666.8255 7600.7423
ENERGY: 2 7916.2985 5742.1035 436.8822 40.9549 -82930.8147 13791.9735 0.0000 0.0000 0.0000 -55002.6022 0.0000 -55002.6022 -55002.6022 0.0000 5432.3391 7370.3167 281259.6748 5432.3391 7370.3167
ENERGY: 3 7878.4431 5735.6817 436.5970 40.9058 -82940.0682 13393.6985 0.0000 0.0000 0.0000 -55454.7421 0.0000 -55454.7421 -55454.7421 0.0000 5016.8834 6964.2358 281259.6748 5016.8834 6964.2358
ENERGY: 4 7804.4690 5723.0900 436.0305 40.8087 -82958.2742 12757.4610 0.0000 0.0000 0.0000 -56196.4151 0.0000 -56196.4151 -56196.4151 0.0000 4345.9939 6314.8795 281259.6748 4345.9939 6314.8795
ENERGY: 5 7663.3786 5698.9262 434.9535 40.6193 -82993.6089 11864.4871 0.0000 0.0000 0.0000 -57291.2443 0.0000 -57291.2443 -57291.2443 0.0000 3384.6148 5398.6693 281259.6748 3384.6148 5398.6693
ENERGY: 6 7408.1034 5654.2007 433.5232 40.2601 -83060.8871 10739.2295 0.0000 0.0000 0.0000 -58785.5701 0.0000 -58785.5701 -58785.5701 0.0000 2131.4370 4224.4109 281259.6748 2131.4370 4224.4109
- Wouldn't it be great to run NAMD a bit faster ? Say, approximately three times faster ? Isn't it a pity to have pc08 and pc10 filtering the dust in the terminal room ? While you are at /work/tmp/my_tests/ create a file with the name NAMD.sh containing the following :
- This script is a parallel NAMD job submission script for the Sun Grid Engine. All you have to do is : qsub NAMD.sh (from the /work/tmp/my_tests/ directory). You should see a message containing your job's ID.
- Type qstat : you should see your job and its status changing from qw to t to r.
- Use mosmon to confirm that the previously idle machines are indeed doing something useful.
- Use qmon to review cluster usage, and to see your job in Job Control/Running Jobs.
- tail -f LOG should show you the growing log file from your job.
- Type mknamdplot LOG and then click Draw to see a collection of graphs showing the evolution of quantities like the system's total energy, pressure, temperature, bond and angle energies, … :
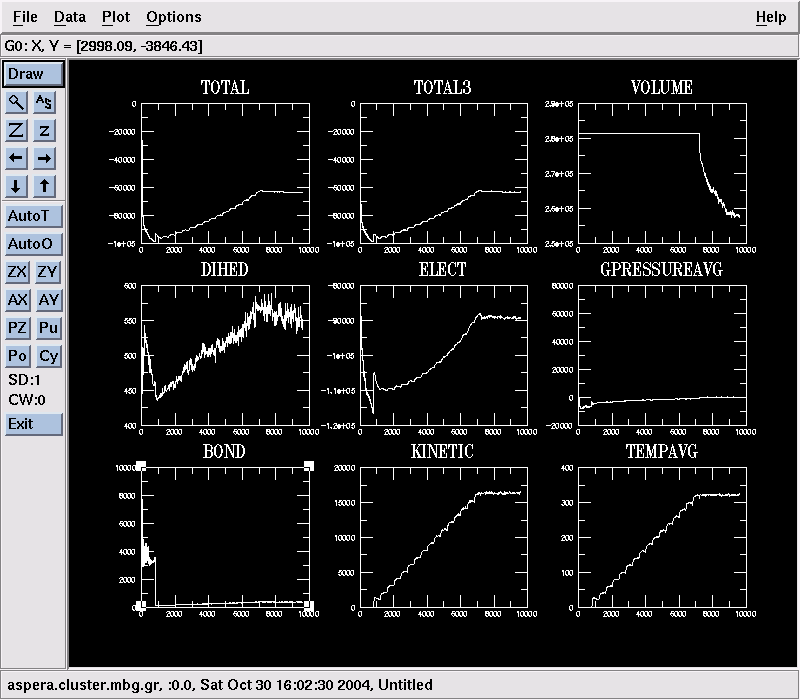 Click Exit to exit.
Click Exit to exit. - The heating-up NAMD job will take 2-3 hours to finish. Let it do so. But if you'd rather not : qdel JobID will kill your job (or you can use the qmon GUI).
- When the job is done review the files that have been created in /work/tmp/my_tests/output/. You should see something like
-rw-rw-r-- 1 glykos glykos 2029503 Oct 30 15:19 equil_ca.coor
-rw-rw-r-- 1 glykos glykos 2029502 Oct 30 15:19 equil_ca.vel
-rw-rw-r-- 1 glykos glykos 222 Oct 30 15:19 equil_ca.xsc
-rw-rw-r-- 1 glykos glykos 2029503 Oct 30 15:16 heat_ca.coor
-rw-rw-r-- 1 glykos glykos 2029503 Oct 30 15:13 heat_ca.coor.BAK
-rw-rw-r-- 1 glykos glykos 2029502 Oct 30 15:16 heat_ca.vel
-rw-rw-r-- 1 glykos glykos 2029502 Oct 30 15:13 heat_ca.vel.BAK
-rw-rw-r-- 1 glykos glykos 153 Oct 30 15:16 heat_ca.xsc
-rw-rw-r-- 1 glykos glykos 153 Oct 30 15:13 heat_ca.xsc.BAK
-rw-rw-r-- 1 glykos glykos 2029503 Oct 30 15:36 heat_out.coor
-rw-r--r-- 1 glykos glykos 14613396 Oct 30 15:36 heat_out.dcd
-rw-rw-r-- 1 glykos glykos 2029502 Oct 30 15:36 heat_out.vel
-rw-rw-r-- 1 glykos glykos 227 Oct 30 15:36 heat_out.xsc
-rw-rw-r-- 1 glykos glykos 3364 Oct 30 15:36 heat_out.xst
-rw-rw-r-- 1 glykos glykos 2029502 Oct 30 14:25 min_all.coor
-rw-rw-r-- 1 glykos glykos 2029501 Oct 30 14:25 min_all.vel
-rw-rw-r-- 1 glykos glykos 152 Oct 30 14:25 min_all.xsc
-rw-rw-r-- 1 glykos glykos 2029502 Oct 30 14:19 min_fix.coor
-rw-rw-r-- 1 glykos glykos 2029501 Oct 30 14:19 min_fix.vel
-rw-rw-r-- 1 glykos glykos 152 Oct 30 14:19 min_fix.xsc
-rw-rw-r-- 1 glykos glykos 608836 Oct 30 15:31 restart.coor
-rw-rw-r-- 1 glykos glykos 608836 Oct 30 15:22 restart.coor.old
-rw-rw-r-- 1 glykos glykos 608836 Oct 30 15:31 restart.vel
-rw-rw-r-- 1 glykos glykos 608836 Oct 30 15:22 restart.vel.old
-rw-rw-r-- 1 glykos glykos 229 Oct 30 15:31 restart.xsc
-rw-rw-r-- 1 glykos glykos 228 Oct 30 15:22 restart.xsc.old
- Play time : from the output/ directory give vmd ../ionized.psf heat_out.dcd
- You can now start preparing the equilibration-production run :
- cd /work/tmp/my_tests/
- mkdir heat
- mv * heat/
- mkdir equi
- cd heat/output
- cp heat_out.coor heat_out.vel heat_out.xsc ../../equi/
- cd ..
- cp ionized.p* par_all27_prot_na.inp ../equi/
- cd ../equi
- mkdir output
- Create a NAMD equilibration script with the name equi.namd containing the following :
- Prepare a submission script for SGE, say NAMD.sh :
- Run it : qsub NAMD.sh
- Enjoy.
This graph shows (in real time) the CPU load for each of the nodes that are currently part of the cluster. The horizontal axis shows the various nodes, the vertical axis shows the corresponding load. If all machines are working properly and they are all running the openMosix linux kernel you should see at least 18 nodes (numbered 25700-25716, plus another node with ID 25854). In this case all cluster nodes are running a proper operating system (and can be seen in the mosmon output) and three of these (25708, 25709 and 25710 corresponding to pc08, pc09 and pc10) are idle. So, nodes pc08, pc09 and pc10 are good candidates for performing your tests.
Select "Queue control". This should pop-up the corresponding window :
Note the small green boxes (indicating that a job is running) on all nodes except pc08, pc09 and pc10. So, it does look like we could use these three machines for the tests. Press 'Done' on the queue control window and then exit in the qmon window.
Click Exit to exit.
![[Home]](/wiki.png)